What does the code do?¶
The Goal of waterpy is to eatimate the potential around cluster.
The Masterfunction: Waterpy¶
That’s how waterpy works:
Import python modules
Sort imported values
Calculate basic values
Start loop:
- Timestep
- Protonation of Ions
- Activity Coefficient
- Supersaturation
- dG per Cluster
- New Cluster J
- Add ion to cluster
- Remove ion to cluster
Modules¶
This function controls all modules.
It imports settings and data and starts the nucleation. Also the logging is done here. @author: ralf.ruehl
Textin - files¶
Reads input text files, comma seperated values. The following files are required:
| file group | content |
|---|---|
| ions... | describes the ion in solution |
| solution | describes the solution |
| conc gives | concentr of the ions in solution |
| settings-capo | salt specific parameters like SP.. |
Ions¶
| filenames | |
|---|---|
| ion_po4_0.txt | ion_h3o_0.txt |
| ion_hpo4_0.txt | ion_h2o_0.txt |
| ion_h2po4_0.txt | ion_oh_0.txt |
| ion_h3po4_0.txt | ion_ca_0.txt |
Every Ion gets a single file. This fils contains a list with following entries:
| ionname | charge | protons | radius | pKs | pKs | pKs | n of pKs |
|---|---|---|---|---|---|---|---|
| [caplet] | [eV] | [n] | [pm] | [] | [] | [] | [n] |
| PO4 | -3 | 3 | 362 | 12.32 | 7.21 | 2.16 | 3 |
| Ca | +2 | 0 | 123 | 0 | |||
| H3O | +1 | 0 | 264 | 0 |
The pKs is given for the ion and the given ion +H. e.g. in the po file the pHs is given for the reaction:
(1)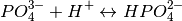
water - solution¶
filename: settings-solution.txt
| pH | volume | temperature | pressure | loop | time |
|---|---|---|---|---|---|
| [] | [L] | [K] | [bar] | [] | [] |
| 7 | 1 | 298 | 1 | 0 | 0 |
Ionpusher¶
The Ionpusher class collects functions which calculate energies in the loop.
supportive functions to calc the cluster
Created on Sat Oct 29 23:48:22 2011
@author: - ralff.ruehl
- ionpusher.ion_pusher.addion(timestep, rand)¶
decides whether an ion should be added based on the ration
- ionpusher.ion_pusher.cluster(cluster, solconc, layer, settings)¶
calculates the clusters behaviour
e.g. ions eject/adsorb ions
- ionpusher.ion_pusher.dGrcrit(r, g, dGv)¶
calculate the energy necessary to reach critical nucleation radius
needs the radius of the mineral germ, the surface tension, the energy per vol
- ionpusher.ion_pusher.delion(cluster, ratio)¶
decides whether an ion should be removed
- ionpusher.ion_pusher.deltheion(cluster, ratio)¶
decids which ions to remove
needs cluster and environment
- ionpusher.ion_pusher.gauss(x)¶
gauss formula, for statistical occations
- ionpusher.ion_pusher.getnewion(cluster, ratio)¶
decides whether the next ion is an anion or cation
requires list with clusterions and the disired ratio
- ionpusher.ion_pusher.layer(layer, solconc, cluster, settings)¶
calculate the layers behaviour
e.g. take/eject H
- ionpusher.ion_pusher.makeevent()¶
creats an empty list of possible events
lists the most important possible events
- ionpusher.ion_pusher.mwg(ions)¶
returns K
needs concentrations
- ionpusher.ion_pusher.newJ(temp=298.0, dGn=-1.66053878316e-24, supersat=0.1, factor=1)¶
calculate number of new germs
needs temp [K], conc, dGn, straight from physical chemistry, A supersaturation factor arbitrary value to influence J
- ionpusher.ion_pusher.newactcoeff(A, zp, zn, I, B, C)¶
calc the current activity coefficient
needs ion, ....
- ionpusher.ion_pusher.protonation(pH=7, N=3, zB=-3, Ct=1)¶
calc the protonation at given pH
N numbers of protons zB = -3 # charge deprot anion Ct = concentration mol / L returns the conc distr of...
- ionpusher.ion_pusher.solsupersat(solconc, Ksp1)¶
calculates the supersaturation of the solution
requires concentrations and solubility products, returns the supersaturation
- ionpusher.ion_pusher.surface(r, ext)¶
calcs surface e dep. from r and an external info ext.
returns the factor?!?!
- ionpusher.ion_pusher.surfacee(cluster, surfacee, time)¶
calc surfaceenergy for a given clustersize and chem. environment
requires cluster diameter, last energy, time interval
- ionpusher.ion_pusher.timestep(lasttimestep=23, event=1, lasttimestepsize=1, temp=293)¶
calculate time interval for this loop
increase: if nothing has happend, or if far away from threshold decrease: if close to threshold or s.th. has happend
the size may vary, so longer timeintervals may be calculated
- ionpusher.ion_pusher.vol(r, bind)¶
gives bindingenergy dep. on clustersize
requires the radius and ...?!?!?
Ioncluster¶
An array of numbers, which represent the ions in a cluster.
Defining all properties of an Ioncluster
Created on Tue Nov 29 17:08:05 2011
@author: rr
- class ionpusher.Ioncluster.Ioncluster¶
Collection of Methods in the Ioncluster
depending on Ions
- chemchange()¶
changes inside the cluster like deprotonation
mainly deprotonation?
- delion()¶
which ion leaves?
find the ion and delete it from cluster
- ions()¶
determines the ions present in the cluster
may vary
- newion(newion)¶
which kind of ion comes?
- noOfIons(ion)¶
count a special Ione in the cluster
returns no of this ion
- surface()¶
gives surface area from cluster
w/o hy shell
Poly Peptide¶
A function serving as an ion pit or source, depending on ab initio output or literature data.
Describes the adsorption of Ions to an Aminoacid String
Created on Tue Jan 17 08:27:55 2012
@author: ralf
- class ionpusher.PolyPeptide.PolyPeptide¶
create a polypeptide string
VesselWall¶
A function serving as an ion pit or source, depending on the surface.
defines the properties of the wall of a vessel to describe the adsorption of ions to the wall of devices
Created on Tue Jan 17 08:32:00 2012
@author: ralf
Textin - Modules¶
Settings and starting scenarios are defind in configuration files and read by the Textin Modules.
Created on Sun Nov 13 16:34:45 2011
@author: -ralf.ruehl read input data from plain text files
- textin.plain.checkpath(path, pathname)¶
checks if the given path for input data is valid
later: further testing for valid input returns true or false
- textin.plain.getiondata(filename, path)¶
reads a file with ion data for the calculations
requires a filename, and path to input dir, returns a list
- textin.plain.getplotdata(filename, path)¶
which of the calculated data shall be plotted?
read from configuration file
- textin.plain.getsettings(filename, path)¶
reads a file with settings for the calculations
requires a filename and path, returns a list
- textin.plain.getsolconc(filename, path)¶
reads a file with the concentration of the ions in the solution
requires a filename and path, returns a list
- textin.plain.getsolprop(filename, path)¶
reads a file with properties of the solution for the calculations
requires a filename, and path, returns a list
- textin.plain.striplist(l)¶
removes whitespaces and from a list
Text out - Modules¶
Severals output options shell be programmed: 1. plain text for arbitary reuse of the data, e.g. in gnuplot #. csv for spreadsheet imports #. export for special molecularmecanics calculations